Marc de Manuel

Postdoctoral Fellow
Columbia University
Email me
Twitter
Bluesky
About me
I am a computational biologist working in the lab of Molly Przeworski at Columbia University. Every biological adaptation emerges from an accidental alteration to the genome that, through genetic shuffling and the interplay of drift and selection, eventually establishes itself in a population. My current research focuses on understanding two fundamental mechanisms in this evolutionary process: mutation and recombination.
You can find a list of publications at my Google scholar profile.
The determinants of germline mutations in mammals
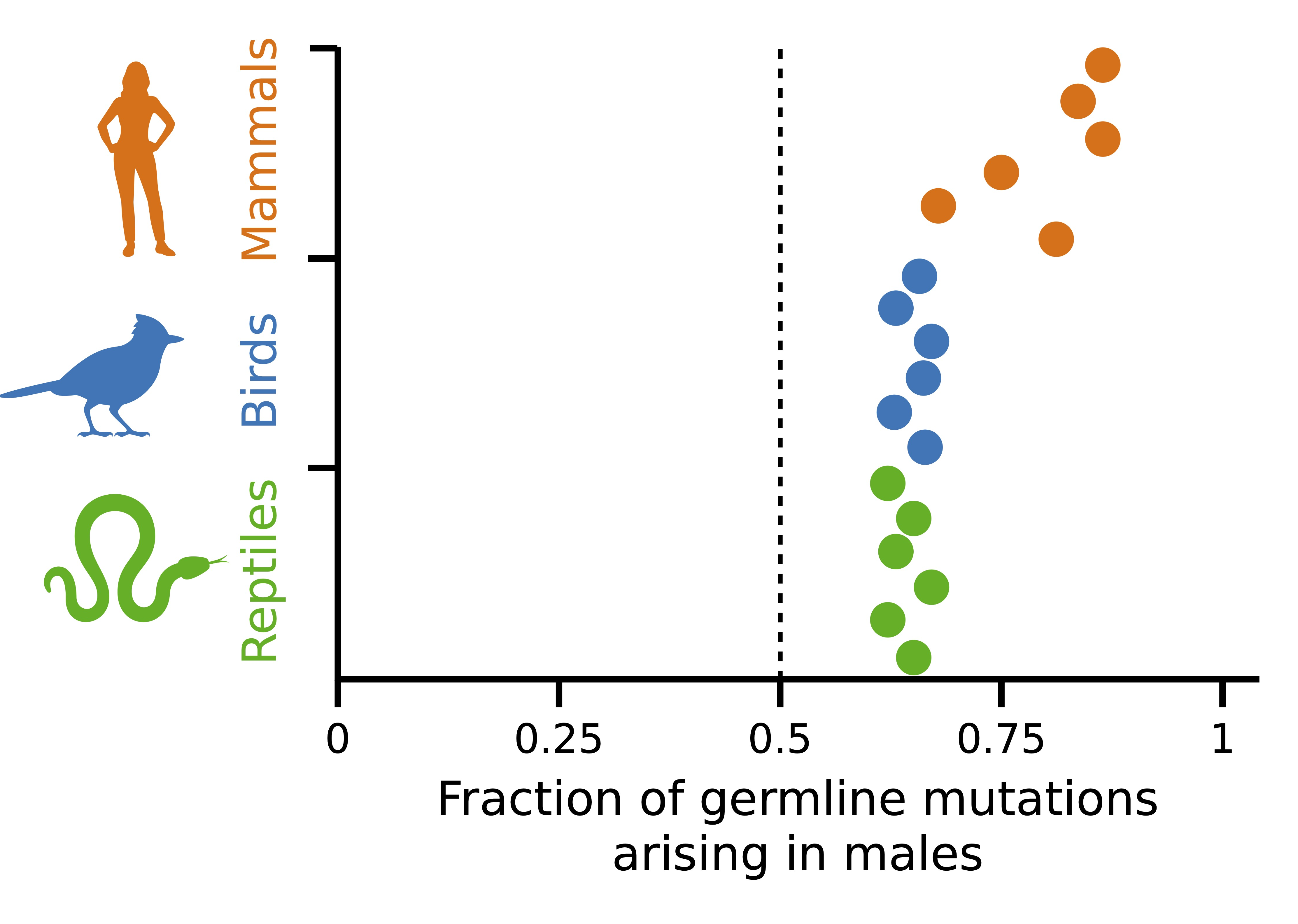
In humans, germline mutations arise more often in males than in females, a phenomenon termed “male mutation bias.” In a recent publication, Felix Wu and I leveraged differences in substitution rates in sex chromosomes vs. chromosomes to show that the bias also occurs in 42 species of amniotes. We provide a novel model to explain the phenomenon in light of damage-induced mutation, helping to reconcile with recent evidence suggesting replication errors are not the main source of germline mutation. You can find the code to reproduce our findings in this link.
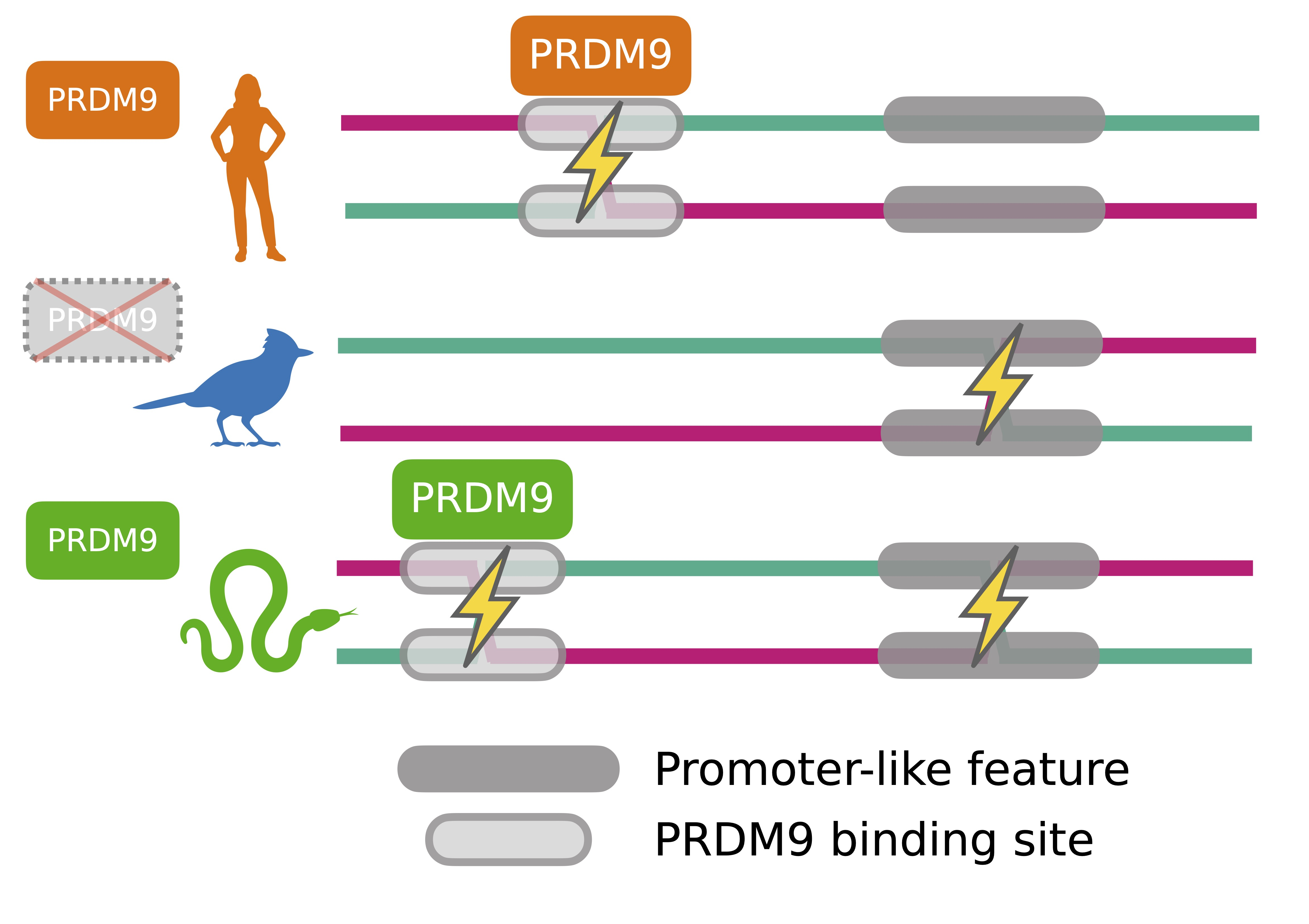
The localization of recombination across vertebrates
Until now, it was believed that the location of recombination in vertebrates was determined by one of two mechanisms: one associated with the protein PRDM9 (used by, e.g., humans and mice) and another by promoter-like features (e.g., used by birds and canids). By studying patterns of linkage disequilibrium and crossover events in pedigrees of corn snakes, Carla Hoge and I show that these features are not mutually-exclusive, and instead reflect a tug of war between competing signals that has more lopsided outcomes in some species than others. Please check the preprint and code to learn more.